Change the present working directory
change the present working directory to the location where the NetCDF file should be written (for example to the location of the m-file in case you are running a script). You can also use a directory called 'processed'.
cd(fileparts(mfilename('fullpath')))
Get data into memory
This array contains altitude data for 1 transect on 3 different timesteps. Each row is a different year of measurement. In this case data is typed in, usually you have some raw input files which you want to read.
data.series = [7.62 7.49 8.26 7.91 7.72 6.03 5.41 5.26 4.82 4.66 4.54 4.17 3.89 3.64 3.55 3.24 3.01 2.72 2.38 2.18 1.96 1.77 1.69 1.53 1.1 0.72 0.3 0.06 0.05 0.01 0.03 -0.06 0 0.09 0.25 0.41 0.37 0.32 0.32 0.33 0.17 0 -0.21 -0.46 -0.67 -0.87 -0.9 -0.92 -0.955 -0.99 -0.96 -0.93 -1.015 -1.1 -1.17 -1.24 -1.39 -1.54 -1.685 -1.83 -1.98 -2.13 -2.3 -2.47 -2.63 -2.79 -2.92 -3.05 -3.175 -3.3 -3.385 -3.47 -3.505 -3.54 -3.49 -3.44 -3.09 -2.74 -2.3 -1.86 -1.905 -1.95 -2.035 -2.12 -2.225 -2.33 -2.435 -2.54 -2.67 -2.8 -2.92 -3.04 -3.145 -3.25 -3.38 -3.51 -3.7 -3.89 -4.08 -4.27 -4.425 -4.58 -4.715 -4.85 -4.945 -5.04 -5.145 -5.25 -5.375 -5.5 -5.73 -5.96 -6.095 -6.23 -6.135 -6.04 -5.925 -5.81 -5.845 -5.88 -5.91 -5.94 -5.99 -6.04 -6.125 -6.21 -6.315 -6.42 -6.485 -6.55 -6.505 -6.46 -6.4 -6.34 -6.325 -6.31 -6.255 -6.2 -6.095 -5.99 -5.845 -5.7 -5.525 -5.35 -5.22 -5.09 -4.995 -4.9 -4.86 -4.82 -4.805 -4.79 -4.805 -4.82 -4.835 -4.85 -4.905 -4.96 -4.99 -5.02 -5.055 -5.09 -5.2 -5.31 -5.375 -5.44 -5.515 -5.59 -5.66 -5.73 -5.83 -5.93 -6.01 -6.09 -6.18 -6.27 -6.365 -6.46 -6.525 -6.59 -6.69 -6.79 -6.865 -6.94 -7.045 -7.15 -7.23 -7.31 -7.39 -7.47 -7.545 -7.62 -7.705 -7.79 -7.845 -7.9 -7.99 -8.08
7.64 7.49 7.95 8.54 8.34 7.54 6.62 6.03 5.28 4.67 4.4 4.18 3.69 3.24 2.9 2.69 2.39 2.05 1.89 1.68 1.5 1.36 1.18 1.03 0.89 0.74 0.6 0.49 0.39 0.27 0.23 0.13 0 -0.11 -0.22 -0.32 -0.38 -0.45 -0.5 -0.51 -0.41 -0.55 -0.55 -0.57 -0.6 -0.63 -0.855 -1.08 -1.365 -1.65 -1.845 -2.04 -2.235 -2.43 -2.53 -2.63 -2.755 -2.88 -3.01 -3.14 -3.195 -3.25 -2.975 -2.7 -2.34 -1.98 -1.73 -1.48 -1.495 -1.51 -1.68 -1.85 -2.335 -2.82 -3.205 -3.59 -3.595 -3.6 -3.78 -3.96 -4.005 -4.05 -4.11 -4.17 -4.32 -4.47 -4.62 -4.77 -4.73 -4.69 -4.77 -4.85 -4.535 -4.22 -3.88 -3.54 -3.295 -3.05 -2.975 -2.9 -2.875 -2.85 -2.865 -2.88 -2.935 -2.99 -3.07 -3.15 -3.38 -3.61 -3.92 -4.23 -4.49 -4.75 -4.99 -5.23 -5.43 -5.63 -5.825 -6.02 -6.18 -6.34 -6.515 -6.69 -6.63 -6.57 -6.575 -6.58 -6.61 -6.64 -6.605 -6.57 -6.66 -6.75 -6.7 -6.65 -6.665 -6.68 -6.54 -6.4 -6.305 -6.21 -6.09 -5.97 -5.885 -5.8 -5.755 -5.71 -5.645 -5.58 -5.54 -5.5 -5.47 -5.44 -5.395 -5.35 -5.325 -5.3 -5.29 -5.28 -5.295 -5.31 -5.335 -5.36 -5.39 -5.42 -5.485 -5.55 -5.605 -5.66 -5.745 -5.83 -5.905 -5.98 -6.07 -6.16 -6.23 -6.3 -6.4 -6.5 -6.585 -6.67 -6.755 -6.84 -6.94 -7.04 -7.135 -7.23 -7.31 -7.39 -7.465 -7.54 -7.635 -7.73 -7.8 -7.87 -7.945 -8.02
7.56 7.43 7.95 8.84 8.42 7.7 6.77 6.28 4.9 4.62 3.82 3.1 2.68 2.41 2.14 1.94 1.77 1.6 1.49 1.34 1.23 1.08 0.97 0.99 0.9 0.77 0.65 0.77 0.79 0.63 0.48 0.32 0.16 0 -0.19 -0.54 -0.88 -1.13 -1.27 -1.41 -1.51 -1.61 -1.635 -1.66 -1.62 -1.58 -1.57 -1.56 -1.45 -1.34 -1.15 -0.96 -0.97 -0.98 -1.06 -1.14 -1.24 -1.34 -1.44 -1.54 -1.73 -1.92 -2.155 -2.39 -2.62 -2.85 -3.06 -3.27 -3.6 -3.93 -4.16 -4.39 -4.555 -4.72 -4.77 -4.82 -4.79 -4.76 -4.45 -4.14 -3.71 -3.28 -3.075 -2.87 -2.825 -2.78 -2.79 -2.8 -2.725 -2.65 -2.59 -2.53 -2.52 -2.51 -2.52 -2.53 -2.595 -2.66 -2.81 -2.96 -3.18 -3.4 -3.655 -3.91 -4.125 -4.34 -4.53 -4.72 -4.9 -5.08 -5.23 -5.38 -5.495 -5.61 -5.735 -5.86 -5.955 -6.05 -6.125 -6.2 -6.285 -6.37 -6.43 -6.49 -6.52 -6.55 -6.595 -6.64 -6.65 -6.66 -6.665 -6.67 -6.67 -6.67 -6.66 -6.65 -6.615 -6.58 -6.525 -6.47 -6.395 -6.32 -6.245 -6.17 -6.08 -5.99 -5.915 -5.84 -5.77 -5.7 -5.635 -5.57 -5.53 -5.49 -5.45 -5.41 -5.4 -5.39 -5.39 -5.39 -5.39 -5.39 -5.435 -5.48 -5.505 -5.53 -5.59 -5.65 -5.7 -5.75 -5.815 -5.88 -5.955 -6.03 -6.105 -6.18 -6.255 -6.33 -6.41 -6.49 -6.57 -6.65 -6.74 -6.83 -6.9 -6.97 -7.06 -7.15 -7.22 -7.29 -7.375 -7.46 -7.535 -7.61 -7.695 -7.78 -7.865 -7.95];
For each point cross-shore there is a distance from the 0 point which is roughly at the beach line. Positive is in the direction of the sea.
data.distance = (-65:5:920)';
Data is stored for 3 different years
data.time = (2006:2008)';
Create empty outputfile
Create a new empty netcdf file.
outputfile = 'transect.nc';
nc_create_empty(outputfile);
Add dimensions
nc_add_dimension(outputfile, 'cross_shore', size(data.series,2)) nc_add_dimension(outputfile, 'time', size(data.time,1))
Add variables
One by one each of the variables can now be added to the NetCDF file First we will add the cross-shore variable
s.Name = 'cross_shore'; s.Nctype = nc_double; s.Dimension = {'cross_shore'}; s.Attribute = struct('Name', {'long_name', 'units', 'comment'}, ... 'Value', {'cross-shore coordinate', 'm', 'cross-shore coordinate relative to the 0 point'}); nc_addvar(outputfile, s);
Usually you want to add some coordinate variables like this We have a latitude and longitude coordinate for each cross-shore coordinate. Therefor the dimension is cross_shore. You can also say that latitude and longitudes are a function of the cross-shore distance.
s.Name = 'lat'; s.Nctype = nc_double; s.Dimension = {'cross_shore'}; s.Attribute = struct('Name', {'standard_name', 'units', 'axis'}, 'Value', {'latitude', 'degree_north', 'X'}); nc_addvar(outputfile, s); s.Name = 'lon'; s.Nctype = nc_double; s.Dimension = {'cross_shore'}; s.Attribute = struct('Name', {'standard_name', 'units', 'axis'}, 'Value', {'longitude', 'degree_east', 'Y'}); nc_addvar(outputfile, s);
Time has the dimension time. The time dimension is the number of time steps, while the time variable contains the time value (in this case the year) of the time step. Using a year should generally be avoided because it's a bit ambiguous. You can find some comments on this in the CF convention
s.Name = 'time'; s.Nctype = nc_int; s.Dimension = {'time'}; s.Attribute = struct('Name', {'standard_name', 'units', 'comment', 'axis'}, 'Value', {'time', 'year', 'measurement year', 'T'}); nc_addvar(outputfile, s);
Finally we create a variable to store the altitude. Here we make a reference to the lat and lon coordinates.
s.Name = 'altitude'; s.Nctype = nc_double; s.Dimension = {'time', 'cross_shore'}; s.Attribute = struct('Name', {'standard_name', 'units', 'comment', 'coordinates', '_FillValue'}, ... 'Value', {'surface_altitude', 'm', 'altitude above geoid (NAP)', 'lat lon', -9999}); nc_addvar(outputfile, s);
store values
We can put data into nc file. In this case we can store the whole array in one go. Sometimes an array is too big to fit into memory. You can then loop over variables and use strides.
nc_varput(outputfile, 'altitude', data.series) nc_varput(outputfile, 'time', data.time) nc_varput(outputfile, 'cross_shore', data.distance)
TODO: add the latitude and longitude variable. See the convertjarkus as an example.
dump
To check wether our outputfile looks as expected we can dump the text representation of the header. This is equivalent to doing ncdump -h outputfile on the command line.
nc_dump(outputfile)
netCDF transect.nc {
dimensions:
cross_shore = 198 ;
time = 3 ;
variables:
// Preference 'PRESERVE_FVD': false,
// dimensions consistent with ncBrowse, not with native MATLAB netcdf package.
double cross_shore(cross_shore), shape = [198]
cross_shore:long_name = "cross-shore coordinate"
cross_shore:units = "m"
cross_shore:comment = "cross-shore coordinate relative to the 0 point"
double lat(cross_shore), shape = [198]
lat:standard_name = "latitude"
lat:units = "degree_north"
lat:axis = "X"
double lon(cross_shore), shape = [198]
lon:standard_name = "longitude"
lon:units = "degree_east"
lon:axis = "Y"
int32 time(time), shape = [3]
time:standard_name = "time"
time:units = "year"
time:comment = "measurement year"
time:axis = "T"
double altitude(time,cross_shore), shape = [3 198]
altitude:standard_name = "surface_altitude"
altitude:units = "m"
altitude:comment = "altitude above geoid (NAP)"
altitude:coordinates = "lat lon"
altitude:_FillValue = -9999
}
read values
The file is created and we can now read the values we put in. The variables are returned as arrays.
x = nc_varget(outputfile, 'cross_shore'); y = nc_varget(outputfile, 'time'); z = nc_varget(outputfile, 'altitude');
plot values
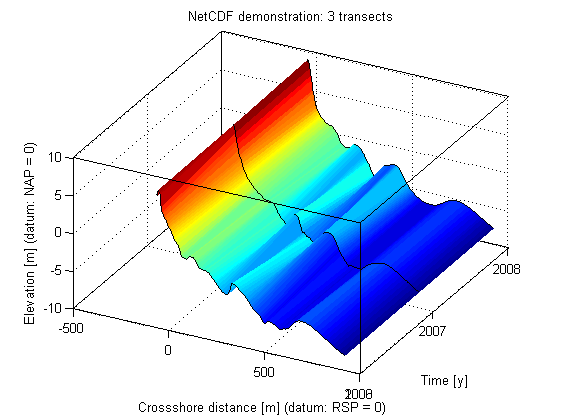
Now let's create some nice plots.
figure(1);clf surface(x, y, z); shading flat hold on; for i = 1:length(y) ph = plot3(x,ones(1,size(z,2)).*double(y(i)),z(i,:)); set(ph,'color', 'k') end set(gca,'ytick', y) box on grid on xlabel('Crossshore distance [m] (datum: RSP = 0)') ylabel('Time [y]') zlabel('Elevation [m] (datum: NAP = 0)') title('NetCDF demonstration: 3 transects') set(gca, 'cameraposition', [4492.56 1996.08 115.897])
Finding out more
If you want to find out more about creating netcdf files, please refer to the following websites: http://cf-pcmdi.llnl.gov CF standard for metadata in netcdf files http://www.unidata.ucar.edu/software/netcdf/ Unidata netcdf pages. You can find the latest versions of the c,c++,fortran and java libraries here as well as mailing lists and a lot of documentation. http://mexcdf.sourceforge.net/ The matlab library (snctools) used in this example. http://www.mathworks.com/access/helpdesk/help/techdoc/ref/netcdf.html the native matlab library available from 2008B